Domain Expertise: What deep learning needs for better COVID-19 detection
27 Sep 2020By now, you’ve probably seen a few, if not many, articles on how deep learning could help detect COVID-19. In particular, convolutional neural networks (CNNs) have been studied as a faster and cheaper alternative to the gold-standard PCR test by just analyzing the patient’s computed tomography (CT) scan. It’s not surprising since CNN is excellent at image recognition; many places have CT scanners rather than COVID-19 testing kits (at least initially).
Despite its success in image recognition tasks such as the ImageNet challenge, can CNN really help doctors detect COVID-19? If it can, how accurately can it do so? It’s well known that CT scans are sensitive but not specific to COVID-19. That is, COVID-19 almost always produces abnormal lung patterns visible from CT scans. However, other pneumonia can create the same abnormal patterns. Can the powerful and sometimes magical CNN tackle this ambiguity issue?
We had a chance to answer these questions ourselves (with my colleague Yuchen and advisor A/P Chen). I’ll walk you through a COVID-19 classifier that we’ve built as an entry to 2020 INFORMS QSR Data Challenge. If you are not familiar with CNN or would like a refresher on the key features of CNNs, I highly recommend reading Convolutional Neural Network: How is it different from the other networks? first. Also, if you’d like to get hands-on, you can get all the code and data from my Github repo.
Key takeaways
- Transfer learning using pre-trained CNN can achieve a really strong baseline performance on COVID-19 classification (85% accuracy).
- However, domain expertise-informed feature engineering and adaptation are required to elevate the CNN (or other ML methods) to a medically convincing level.
What’s the challenge?
COVID-19 pandemic has changed lives around the world. This is the current situation as of 2020/09/26, according to WHO. 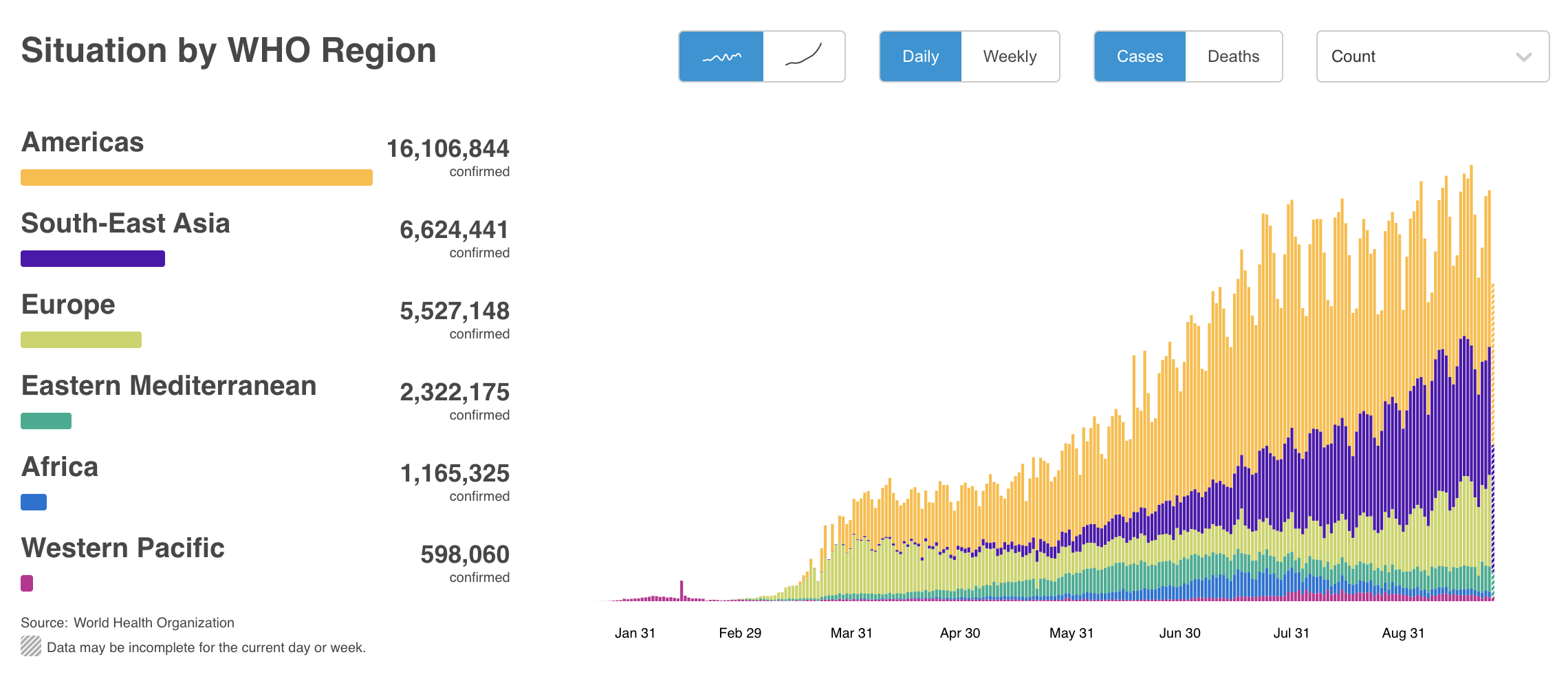
CT scans have been used to screen and diagnose COVID-19, especially in areas where swab test resources are severely lacking. The goal of this data challenge is to diagnose COVID-19 using chest CT scans. Therefore, we need to build a classification model that can classify patients to COVID or NonCOVID based on their chest CT scans, as accurately as possible.
What’s provided?
Relatively even number of COVID and NonCOVID images are provided to train the model. While meta-information of these images is also provided, they will not be provided during testing. The competition also requires that the model’s training with provided data must take less than one hour.
- Training data set
- 251 COVID-19 CT images
- 292 non-COVID-19 CT images
- Meta-information
- e.g., patient information, severity, image caption
All challenge data are taken from a public data set.
Model performance
Let’s first take a look at the result, shall we?
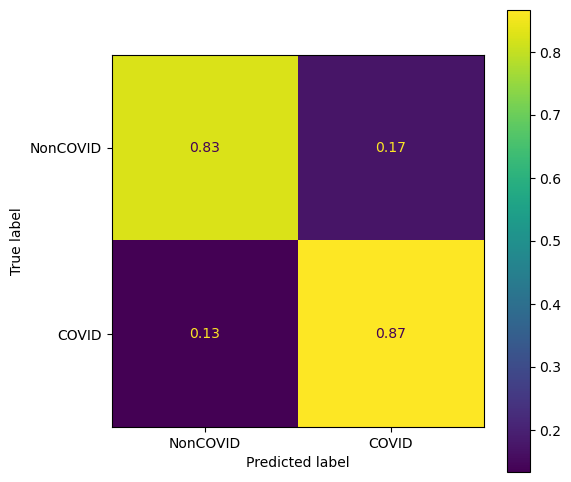
The trained model is evaluated with an independent set of test data. Here you can see the confusion matrix. The overall accuracy is about 85% with slightly better sensitivity than specificity, i.e., true positive rate > true negative rate.
Implementation
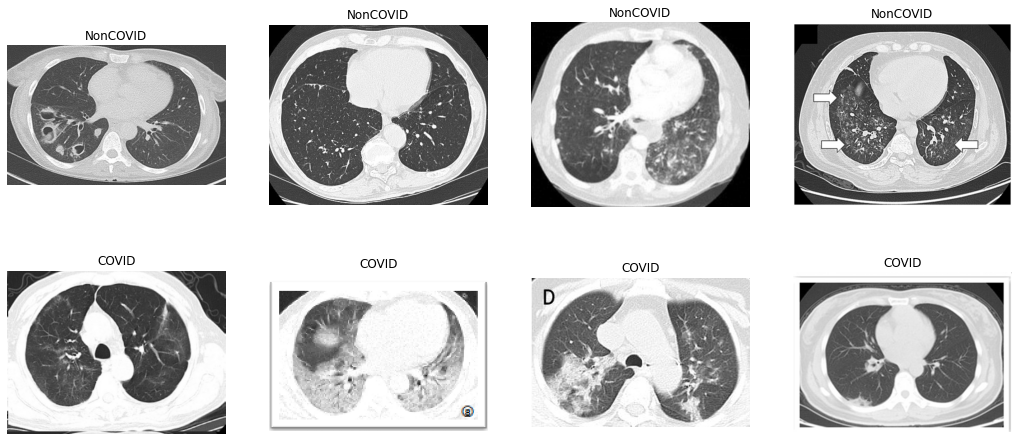
Here are some of the provided NonCOVID and COVID CT scans. It’s important to note that the challenge is to distinguish between COVID and NonCOVID CT scans, rather than COVID and Normal scans. In fact, there may be some NonCOVID CT scans that belong to other pneumonia patients.
Train-validation split
We reserve 20% of the data for validation. Since some consecutive images come from the same patient, they tend to be similar to each other. That is, many of our data are not independent. To prevent data leakage (information of training data spills over to validation data), we keep the original image sequence and hold out the last 20% as the validation set.
After the splitting, we have two pairs of data:
X_train,y_trainX_val,y_val
X is a list of CT scans, and y is a list of binary labels (0 for NonCOVID, 1 for COVID).
Data augmentation
Data augmentation is a common way to include more random variations in the training data. It helps to prevent overfitting. For image-related learning problems, augmentation typically means applying random geometric (e.g., crop, flip, rotate, etc.) and appearance transformation (e.g., contrast, edge filter, Gaussian blur, etc.). Here we use tf.keras.Sequential to create a pipeline in which the input image is randomly transformed through the following operations:
- Random horizontal and vertical flip
- Rotation by a random degree in the range of $[-5\%, 5\%]*2\pi$
- Random zoom in height by $5\%$
- Random translation by $5\%$
- Random contrast adjustment by $5\%$
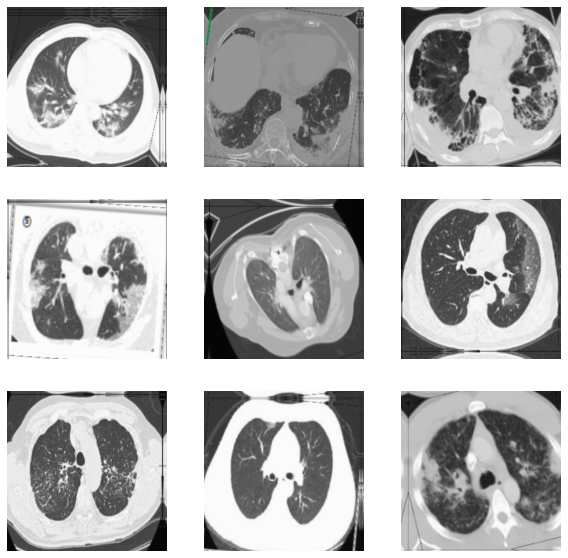
This is how they look after the augmentation.
Using pre-trained CNN as the backbone
We do not build a CNN from scratch. For an image-related problem with only a modest number of training images, it is recommended to use a pre-trained model as the backbone and do transfer learning on that. The chosen model is EfficientNetB0. It belongs to the family of models called EfficientNets proposed by researchers from Google. EfficientNets are among the current state-of-the-art CNNs for computer vision tasks. They
- need a considerably lower number of parameters,
- achieved very high accuracies on ImageNet,
- transferred well to other image classification tasks.
Here’s a performance comparison between EfficientNets and other well-known models: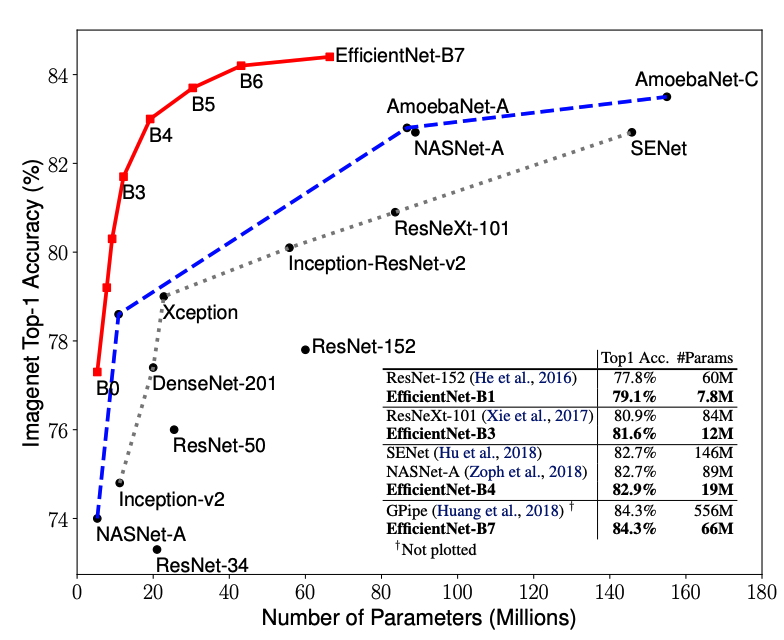
EfficientNets, and other well-known pre-trained models, can be easily loaded from tf.keras.applications. We first import the pre-trained EfficientNetB0 and use it as our model backbone. We remove the original output layer of EfficientNetB0 since it was trained for 1000-class classification. Also, we freeze the model’s weights so that they won’t be updated during the initial training.
# Create a base model from the pre-trained EfficientNetB0
base_model = keras.applications.EfficientNetB0(input_shape=IMG_SHAPE, include_top=False)
base_model.trainable = False
Wrap our model around it
With EfficientNet imported, we can use it to our problem by wrapping our classification model around it. You can think of the EfficientNetB0 as a trained feature extractor. The final model has:
- An input layer
- EfficientNetB0 base model
- An average pooling layer: Pool the information by average operation
- A dropout layer: Set a percentage of inputs to zero
- A classification layer: Output the probability of NonCOVID
We can also use tf.keras.utils.plot_model to visualize our model. 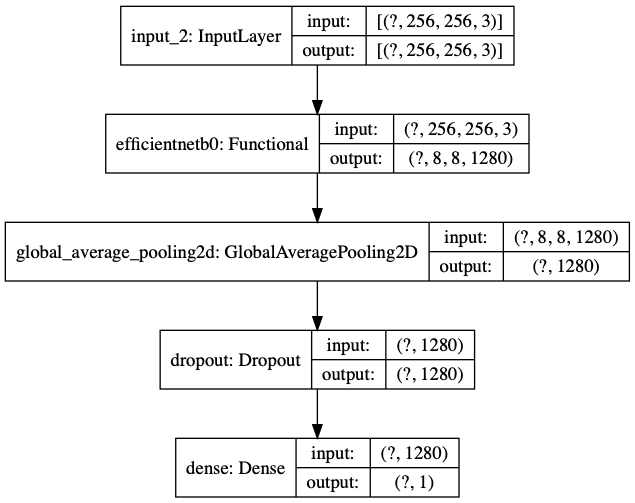
We can see that:
- The
?in input and output shape is a reserved place for the number of samples, which the model does not know yet. - EfficientNetB0 sits right after the input layer.
- The last (classification) layer has an output of dimension 1: The probability for NonCOVID.
Training our model
Public data pre-training: To help EfficientNetB0 adapt to COVID vs NonCOVID image classification, we’ve actually trained our model on another public CT scan data set. The hope is that training the model on CT scans would allow it to learn features specific to our COVID-19 classification task. We will not go into the public data training part, but the procedure is essentially the same as what I will show below.
Transfer learning workflow: We use a typical transfer-learning workflow:
- Phase 1 (Feature extraction): Fix EfficientNetB0’s weights, only update the last classification layer’s weights.
- Phase 2 (Fine tuning): Allow some of EfficientNetB0’ weights to update as well.
You can read more about the workflow here.
Key configurations: We use the following metrics and loss function:
- Metrics: to evaluate model performance
- Binary accuracy
- False and true positives
- False and true negatives
- Loss function: to guide gradient search
- Binary cross-entropy
We use the Adam optimizer, the learning rates is set to [1e-3, 1e-4] and the number of training epochs is set to [10, 30] for the two phases, respectively. The two-phase training is iterated for two times.
Training history: Let’s visualize the training history: 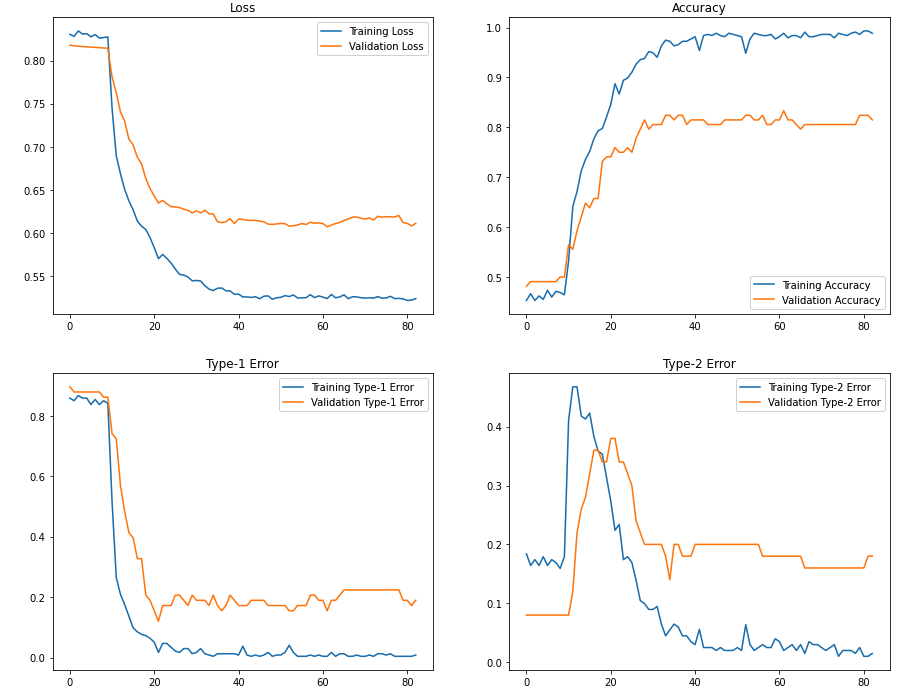
Here you can see that after we’ve allowed some layers of EfficientNets to update (after Epoch 10), we obtain a significant improvement in classification accuracy. The final training and validation accuracy is around 98% and 82%.
How does it perform on test data?
We can obtain a set of test data from the same data repo that contains 105 NonCOVID and 98 COVID images. Let’s see how the trained model performs on them. Here’s a result breakdown using sklearn.metrics.classification_report.
precision recall f1-score support
COVID 0.85 0.83 0.84 98
NonCOVID 0.84 0.87 0.85 105
accuracy 0.85 203
macro avg 0.85 0.85 0.85 203
weighted avg 0.85 0.85 0.85 203
And the ROC curve: 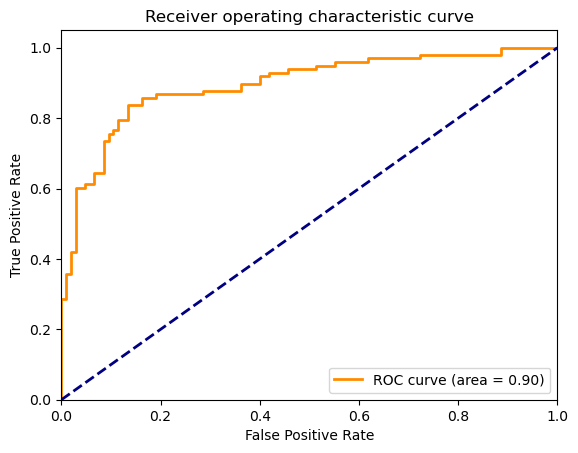
What are correctly and incorrectly classified CT scans?
We can dive into the classification result and see which ones are identified correctly and identified incorrectly. Potential patterns found could be leveraged to help further improve the model.
Could you identify some patterns?
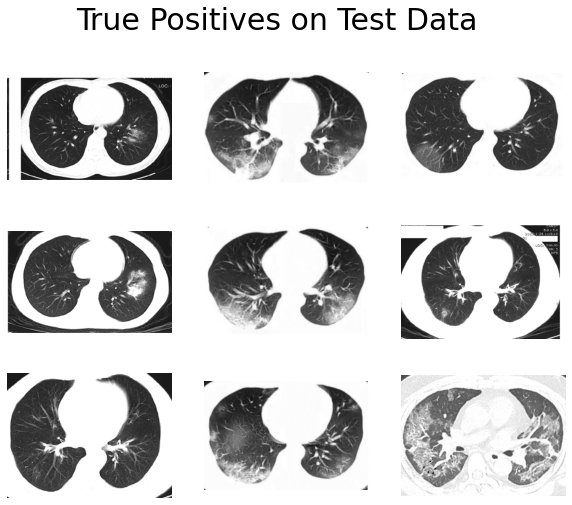
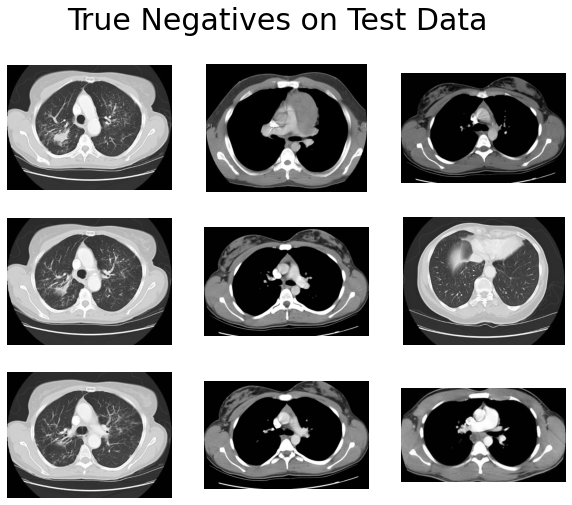
And here are the incorrect ones:
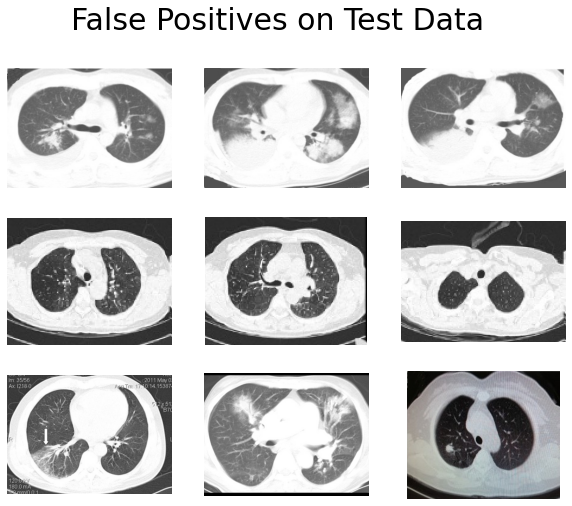
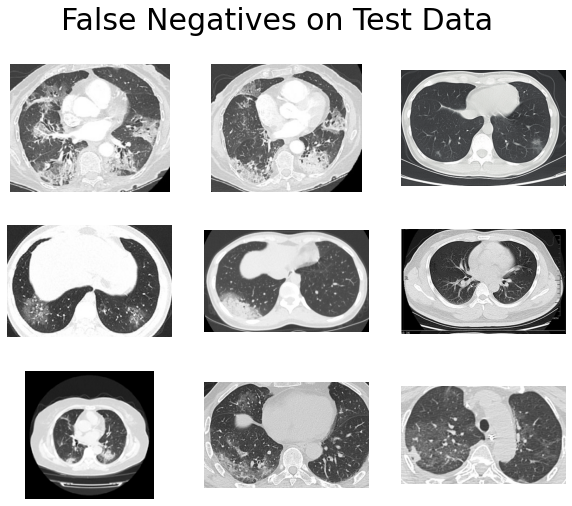
We can probably make several observations here:
- True positives have obvious abnormal patterns, and the lung structures are well-preserved.
- Many of the true negatives have complete black lungs (no abnormal pattern).
- The lung boundaries of many false positives are not clear.
The point is, to a non-medical person like me, many of the COVID and NonCOVID images look the same. The ambiguity is even more severe when some images have unclear lung boundaries. It seems like our CNN is also having trouble distinguishing those images.
Where do we go from here?
From the above results, we can see that a pre-trained CNN can be adapted to achieve a really strong baseline performance. However, there are clear limitations to what a deep learning model (or any other model) alone can achieve. In this case, computer vision researchers and medical experts need to collaborate in a meaningful way so that the end model is both computationally capable and medically reliable.
There are several directions for which we could make the model better:
- Lung segmentation: Process each image and retain only the lung area of the CT scan, for example, see here.
- More sophisticated transfer learning design: For example, see multi-task learning or supervised domain adaptation.
- Ensemble model: This seems like a common belief, especially among Kaggle users, that building an ensemble model would almost always give you an extra few percent accuracy increases.
That’s it for our CNN COVID-19 CT scan classification! Thank you! 👏👏👏